A citable preprint in the bioRxiv describing Bio4j went online yesterday:
It serves (we hope) as a good introduction to what is Bio4j, and what it has to offer; especially so if, for getting a general idea of Bio4j, you would rather read prose than code. If you are using Bio4j for something that you want to publish, citing it is much easier now: all bioRxiv preprints are assigned a DOI. Comments, thoughts, opinions are all more than welcome! We will submit a paper based on this preprint to an open access journal. For completeness, here’s the citation info and the abstract:
Bio4j: a high-performance cloud-enabled graph-based data platform
Pablo Pareja-Tobes, Raquel Tobes, Marina Manrique, Eduardo Pareja, Eduardo Pareja-Tobes
bioRxiv – doi: 10.1101/016758
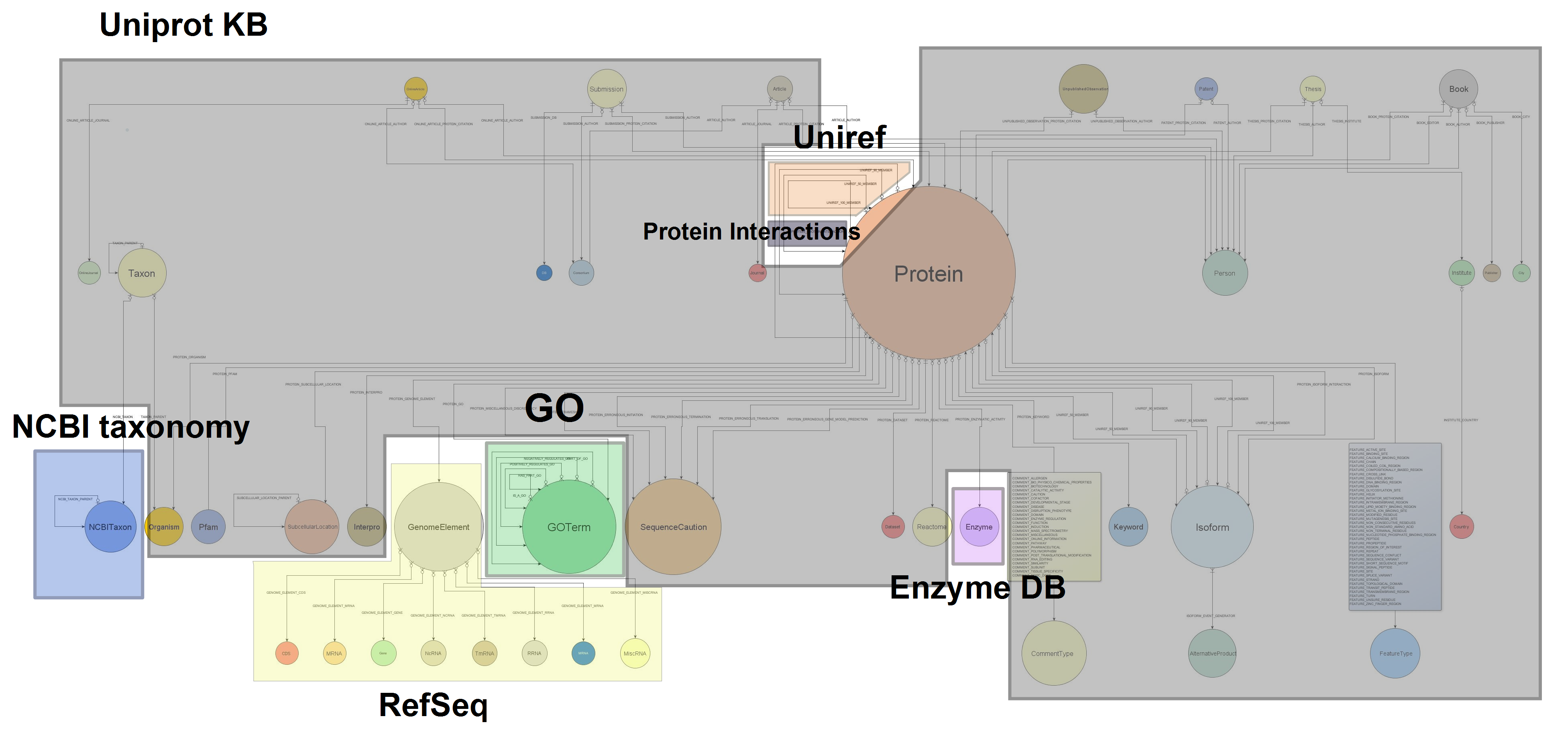
Background. Next Generation Sequencing and other high-throughput technologies have brought a revolution to the bioinformatics landscape, by offering sheer amounts of data about previously unaccessible domains in a cheap and scalable way. However, fast, reproducible, and cost-effective data analysis at such scale remains elusive. A key need for achieving it is being able to access and query the vast amount of publicly available data, specially so in the case of knowledge-intensive, semantically rich data: incredibly valuable information about proteins and their functions, genes, pathways, or all sort of biological knowledge encoded in ontologies remains scattered, semantically and physically fragmented.
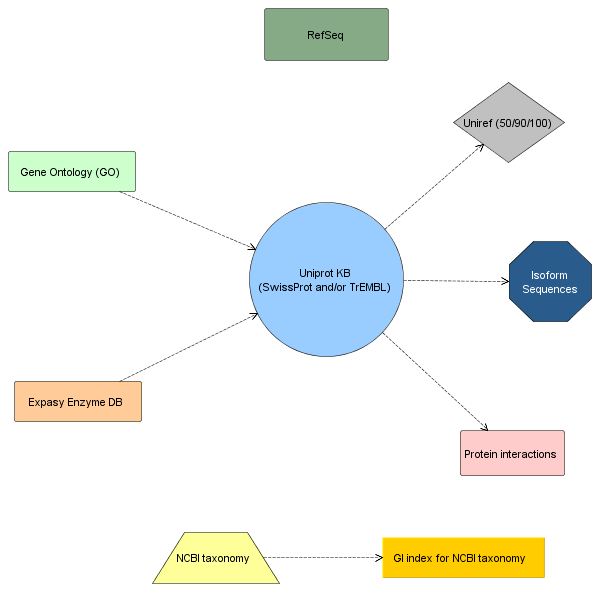
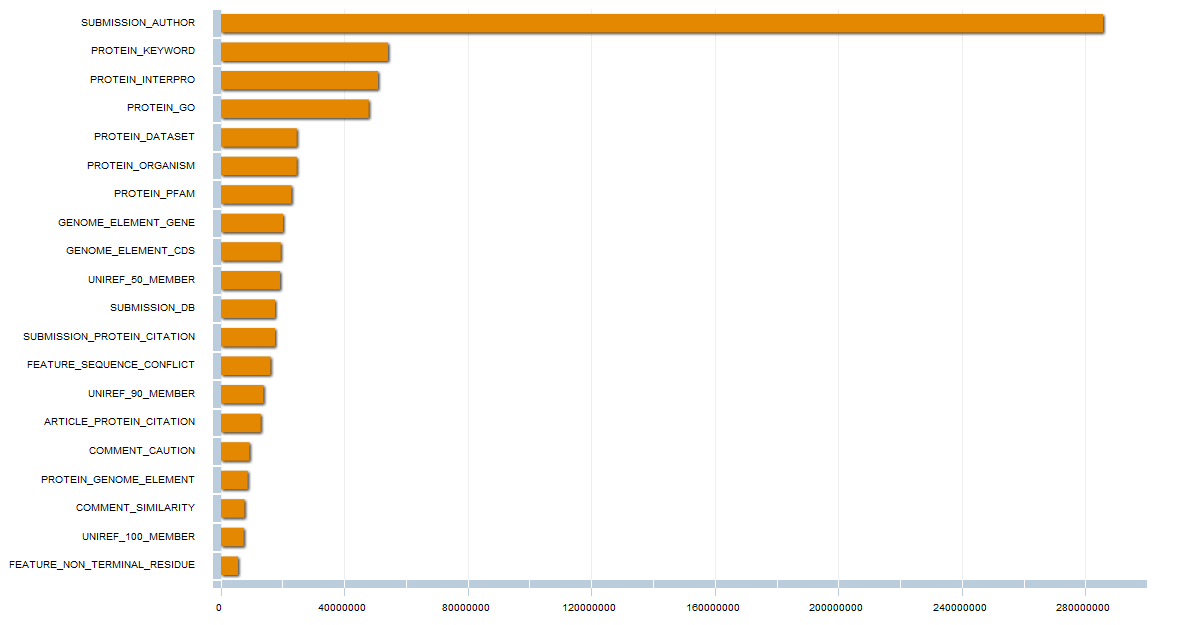
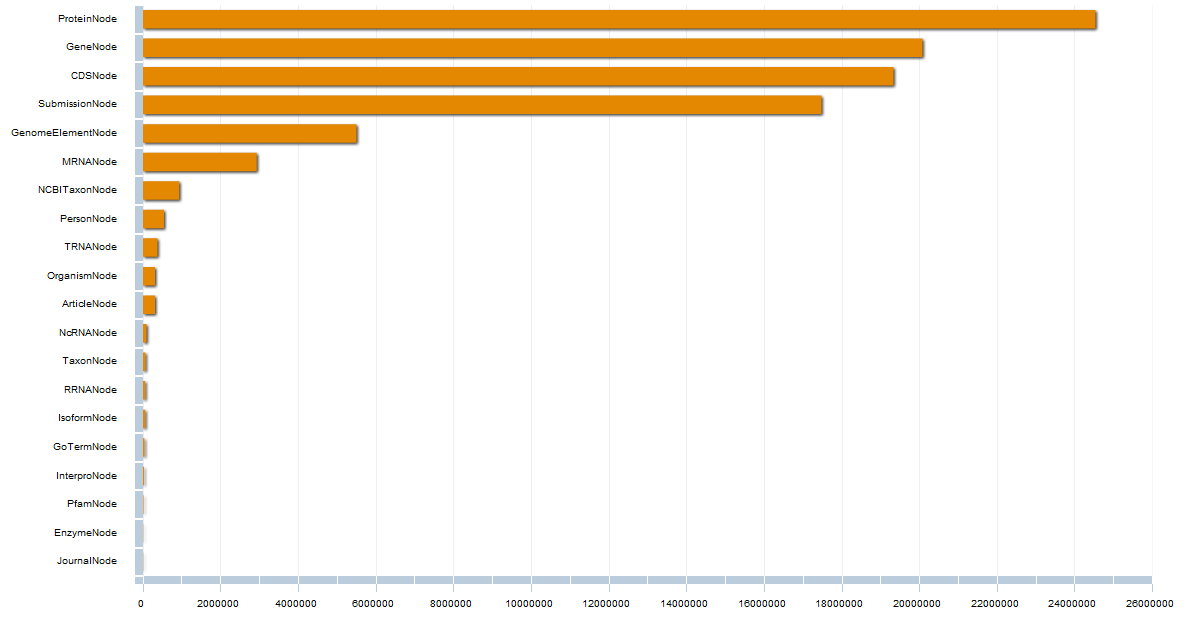
Methods and Results. Guided by this, we have designed and developed Bio4j. It aims to offer a platform for the integration of semantically rich biological data using typed graph models. We have modeled and integrated most publicly available data linked with proteins into a set of interdependent graphs. Data querying is possible through a data model aware Domain Specific Language implemented in Java, letting the user write typed graph traversals over the integrated data. A ready to use cloud-based data distribution, based on the Titan graph database engine is provided; generic data import code can also be used for in-house deployment.
Conclusion. Bio4j represents a unique resource for the current Bioinformatician, providing at once a solution for several key problems: data integration; expressive, high performance data access; and a cost-effective scalable cloud deployment model.







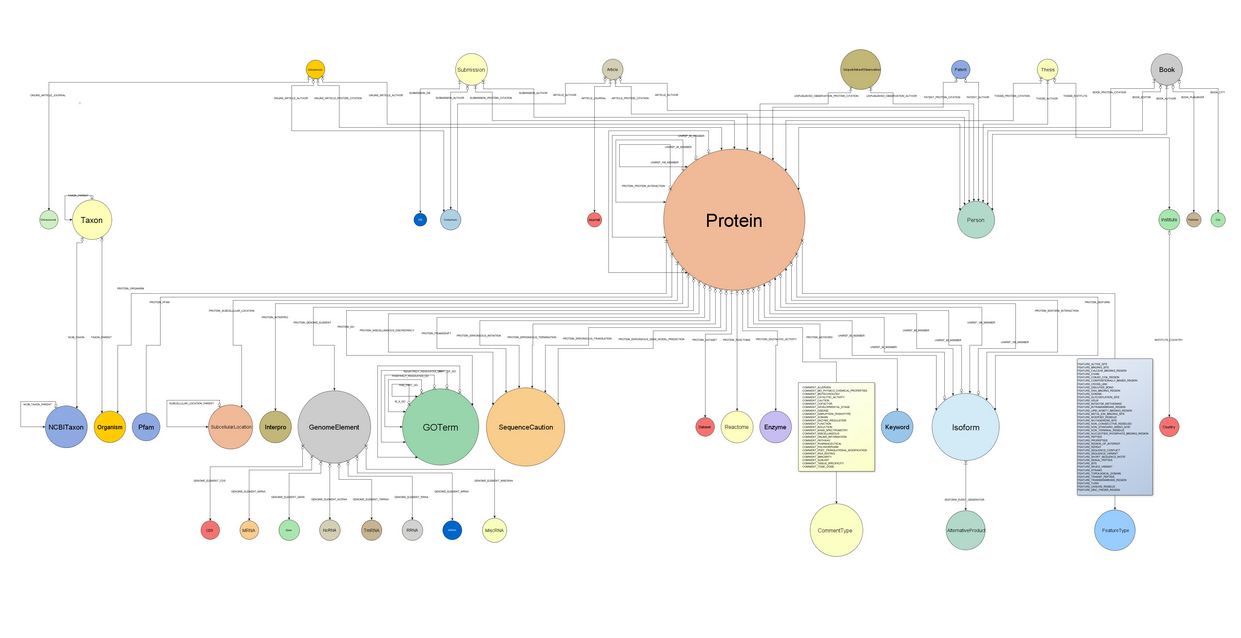
 We have introduced a new level of abstraction for the domain model by decoupling the inner database implementation from the relationships among entities themselves. An interface has been developed for each node and relationship present in the database, including methods to access both the properties of the entity it represents and utility methods that allow to easily navigate to the entities that will be linked to it.
All this can be found under the package com.era7.bioinfo.bio4j.model
We have introduced a new level of abstraction for the domain model by decoupling the inner database implementation from the relationships among entities themselves. An interface has been developed for each node and relationship present in the database, including methods to access both the properties of the entity it represents and utility methods that allow to easily navigate to the entities that will be linked to it.
All this can be found under the package com.era7.bioinfo.bio4j.model Apart from the set of interfaces we’ve developed another layer for the domain model using
Apart from the set of interfaces we’ve developed another layer for the domain model using  After the problems we had with the so called
After the problems we had with the so called