Hi!
I’m happy to announce that the version 0.7 of Bio4j has been released. Check out the wide set of new features, tools and improvements:
Expasy Enzyme database integration
From now on you have the whole Enzyme DB included in Bio4j. For that, both a new node type and relationship type have been created:
- EnzymeNode -ProteinEnzymaticActivityRel (this relationship connects a protein and the respective enzyme nodes associated to it)
All properties found have been incorporated to the enzyme node including:
- ID
- Official name
- Alternate names
- Cofactors
- Comments
- Catalytic activity
- Prosite cross-references
Node type indexing
From now on, every node present in the database has a property nodeType including its type which has been indexed. That way you can now access all nodes belonging to an specific type really easily.
Availability in all Regions
The AWS region you are based in won’t be a problem for using Bio4j anymore. EBS Snapshots have been created in all regions as well as CloudFormation templates have been updated so that they can now be used regardless the region where you want to create the stack.
Only Asia Pacific (Singapore)
ap-southeast-1region is not ready due to ongoing issues from AWS side regarding extremely slow S3 object downloading. Hope we can find a work around for this soon!
New CloudFormation templates
Basic Bio4j instance (updated)
The basic Bio4j instance template has been updated so that now you can use it from all zones. Check out more info about this in the updated blog post
Basic Bio4j REST server
A new template has been developed so that you can easily deploy your Neo4j-Bio4j REST server in less than a minute.
This template is available in the following address:
The steps you should follow to create the stack are really simple. Actually, you can follow as a guide this blog post about the template I created for deploying a general Neo4j server, only one or two parameters vary
Bio4j REST server
Once you get your server running thanks to the useful template I just mentioned before, using Neo4j WebAdmin with Bio4j as source you will be able to:
Explore you database with the Data browser
Using the data browser tab of the Web administration tool you can explore in real-time the contents of Bio4j!
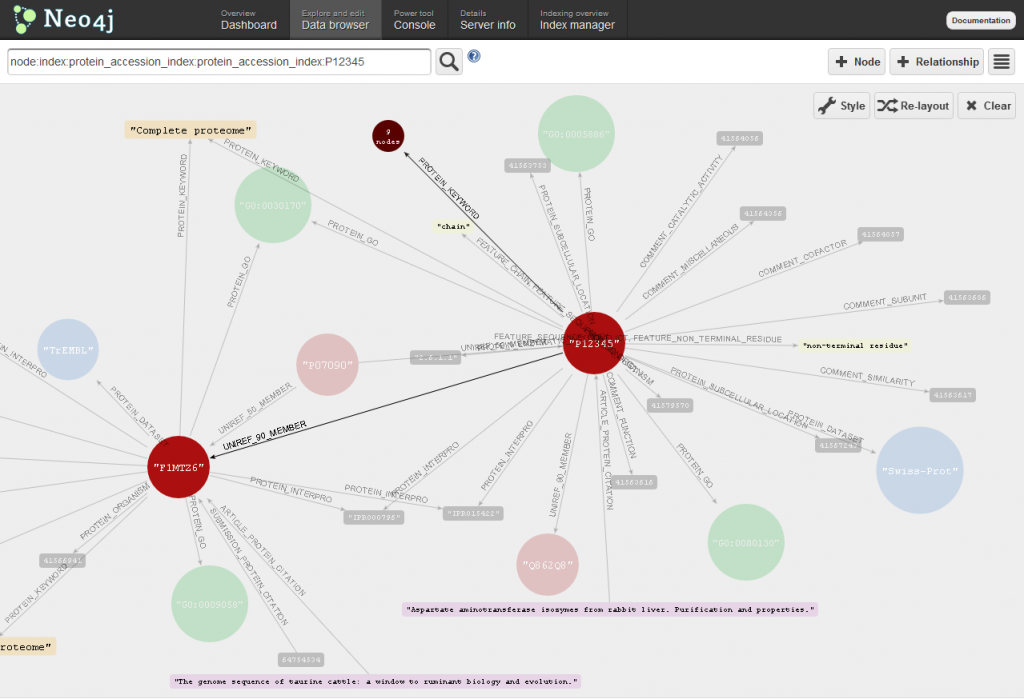
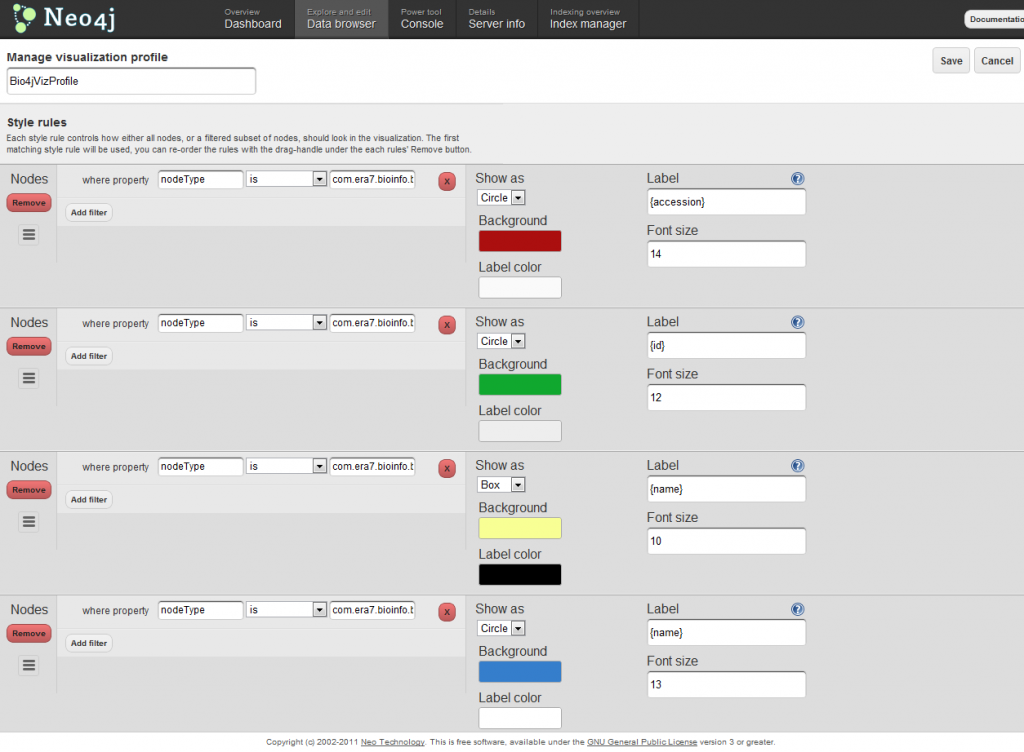
In order to get visualizations like the one shown above, you should make use of visualization profiles. There you can specify different styles associated to customizable rules which can be expressed in terms of the node properties. Here’s a screenshot showing how the visualization profile I used for the visualization above looks like:
Just beware of one thing, the behavior of the tool is such that it does not distinguish between highly connected nodes and more isolated ones. Because of this, clicking nodes such as Trembl dataset node is not advisable unless you want to see it freeze forever -this node has more than 15 million relationships linking it to proteins.
Run queries with Cypher
Cypher what?!
Cypher **is a **declarative language which allows for expressive and efficient querying of the graph store without having to write traversers in code. It focuses on the clarity of expressing what to retrieve from a graph, not how to do it, in contrast to imperative languages like Java, and scripting languages like Gremlin.
A query to retrieve protein interaction circuits of length 3 with proteins belonging to Swiss-Prot dataset (limited to 5 results) would look like this in Cypher:
1 2 3 4 5 | |
If you want to check out more examples of Bio4j + Cypher, check our Bio4j cypher cheat sheet that we will be updating from time to time.
Querying Bio4j with Gremlin
Gremlins? What do they have to do with Bio4j!?
Gremlin is a graph traversal language that can be natively used in various JVM languages - it currently provides native support for Java, Groovy, and Scala. However, it can express in a few lines of code what it would take many, many lines of code in Java to express.
Querying proteins associated to the interpro motif with id IPR023306 in Bio4j with Gremlin would look like this: (limited to 5 results)
1 2 3 4 5 6 7 | |
If you want to check out more examples of Bio4j + Gremlin, check our Bio4j gremlin cheat sheet that we will be updating from time to time.
Bug fixes
- Dataset nodes There was a bug in the importing process which resulted in the creation of a new dataset node everytime a new Uniprot entry was stored. Now everything’s fine!
So that’s all for now! Hope you enjoy all this changes and new features I’ve been working on in the last couple of months. As always, feel free to give any feedback you may have, I’m looking forward to it ;)

