Hi everyone!
After writing this post on December, I’ve been thinking of doing something similar, yet different, using Neo4j Cypher query language.
That’s where I came up with the idea of looking for topological patterns through a large sub-set of the Protein-Protein interactions network included in Bio4j; -rather than focusing in a few proteins selected a priori.
I decided to mine the data in order to find circuits/simple cycles of length 3 where at least one protein is from Swiss-Prot dataset:
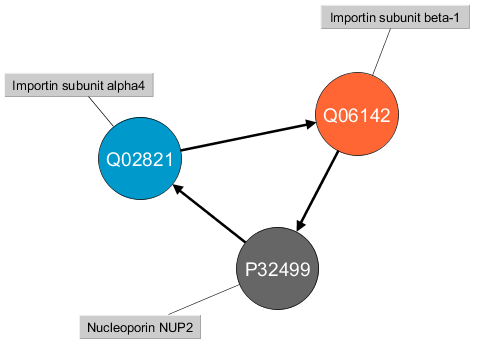
I would like to point out that the direction here is important and these two cycles:
A --> B --> C --> AA --> C --> B --> A
are not the same. Ok, so once this has been said, let’s see how the Cypher query looks like:
1 2 3 4 | |
As you can see it’s really simple and straightforward. In the first two lines we match the proteins from Swiss-Prot dataset for later retrieving the ones which form a 3-length cycle as described before. Once the query has finished, you should be getting something like this:
1 2 3 4 5 6 7 8 9 10 11 | |
As you can see the query took about 17 minutes to be completed in a 100% fresh DB -there was no information cached at all yet; with a m1.large AWS machine -this machine has 7.5GB of RAM.
Not bad, right!?
We have to beware of something though, this query returns cycles such as:
A --> B --> C --> AB --> C --> A --> B
as different cycles when they are actually not. That’s why I developed a simple program to remove these repetitions as well as for fetching some statistics information. After running the program you get two files:
The final circuits found were reduced after performing the filtering to 2226 records.
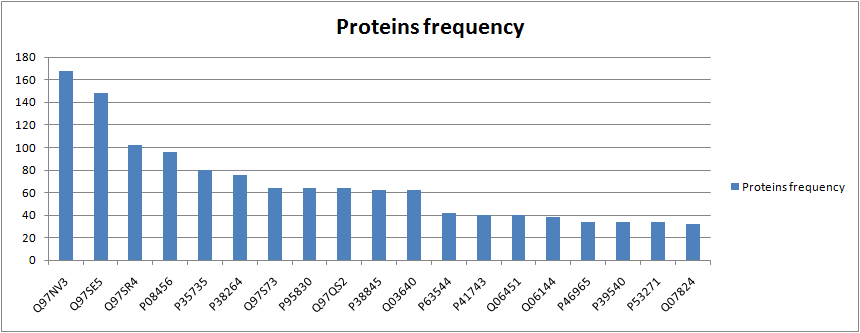
Finally, I also created a really simple chart including the absolute frequency of the first 20 proteins with more occurrences in the cycles that were found.
Well, that’s all for now. Have a good day!
