Hi everyone!
It’s been a few months already since I published the last post but that doesn’t mean that the development process of Bio4j was stopped, but rather, on the contrary, I have been working in the integration of Bio4j with other DB-related projects as well as pipelines and tools. Actually, I’m right now staying in the US for a couple of months working on the implementation and integration of a new database around Bio4j including grasses genomic data as part of a collaboration with the Ohio State University, (I promise to give more details about this and more in next posts).
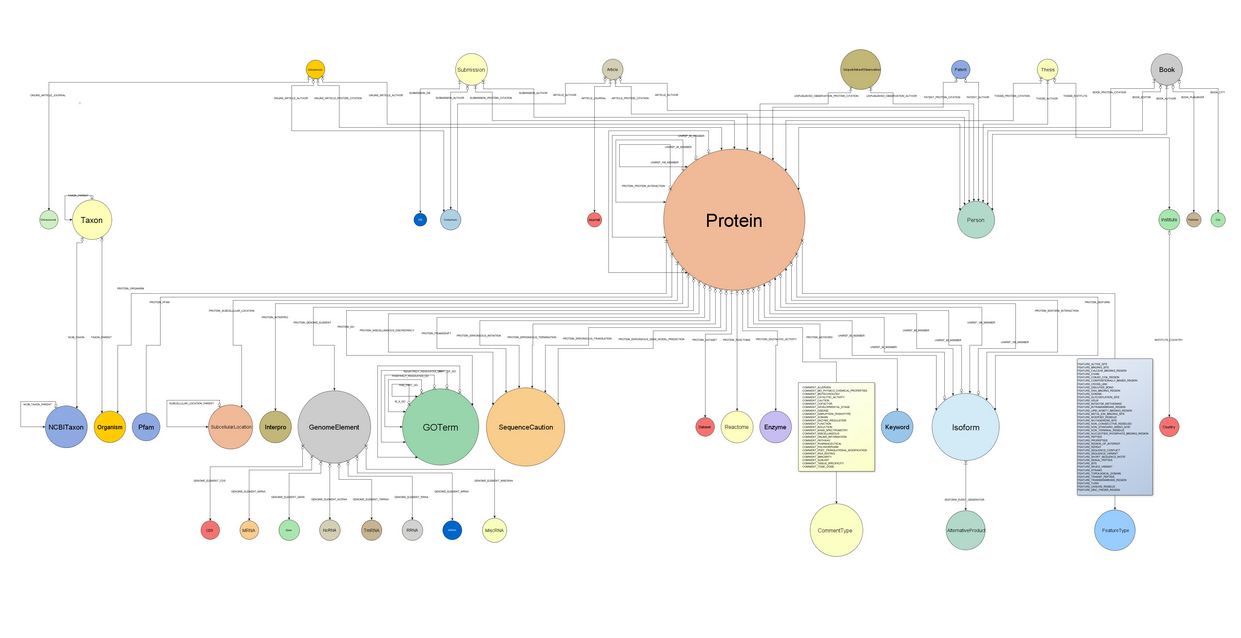
Ok, but let’s get to the point of this post. Even though there already is available a web tool to explore Bio4j data structure (Bio4jExplorer), I was feeling that something else was missing in order to get the big picture of all the data included and how it’s interrelated. So I got to work and created this general domain model including all node types and relationships (also specifying their cardinality).
I didn’t include “auxiliary” relationships linked to the reference node in order to not pollute the schema with relationships that don’t have any semantic meaning but rather indexing purposes. Also, the text included in both boxes represents different relationships all linking the same nodes -specifically Protein with CommentType and FeatureType. I could have drawn them as the rest but then I would have ended up with a hairball instead of a meaningful schema.
As always, any feedback is welcome!